>Clean and Tidy Data
> When you get new data, you have to clean and organize them. Cleaning new data helps you exploring their content and structure
\ Otho Mantegazza _ Dataviz for Scientists _ Part 1.5
Which Dataset is Tidy?
A common practical way to structure (empirical) data.
- Every column is a variable.
- Every row is an observation.
- Every cell is a single value.
- (Every observational unit is in its own table).
Plus: fixed variables should come first, followed by measured variables.
Reference: An Introduction to Tidy Data
Which Dataset is Tidy?

Source: R4DS - Tidy Data
Semantics of (tidy) Data
Always quoting the Tidy Data article:
- A dataset is a collection of values.
- Every value belongs to a variable and an observation.
- A variable contains all values that measure the same underlying attribute (like height, temperature, duration) across units.
- An observation contains all values measured on the same unit (like a person, or a day, or a race) across attributes.
Tools: Tidy data with Tidyr
Tidyr provides functions for:
- Pivoting data.
- Rectangling data.
- Nesting data.
- Combining and splitting columns.
- Make missing values explicit.
Tidy Pangea Data
Remember the dataset from Pangaea?
Tidy Pangea Data
Rows: 128
Columns: 19
$ Event <chr> "SL73BC", "SL73BC", "SL73BC", "SL73BC", "SL…
$ Latitude <dbl> 39.6617, 39.6617, 39.6617, 39.6617, 39.6617…
$ Longitude <dbl> 24.5117, 24.5117, 24.5117, 24.5117, 24.5117…
$ `Elevation [m]` <dbl> -339, -339, -339, -339, -339, -339, -339, -…
$ `Sample label (barite-Sr)` <chr> "SL73-1", "SL73-2", "SL73-3", "SL73-4", "SL…
$ `Samp type` <chr> "non-S1", "non-S1", "non-S1", "S1b", "S1b",…
$ `Depth [m]` <dbl> 0.0045, 0.0465, 0.0665, 0.1215, 0.1765, 0.1…
$ `Age [ka BP]` <dbl> 1.66, 3.13, 4.13, 5.75, 7.30, 7.78, 8.65, 9…
$ `CaCO3 [%]` <dbl> 61.5, 55.1, 53.0, 43.4, 41.8, 42.3, 42.6, 3…
$ `Ba [µg/g] (Leachate)` <dbl> 72.6, 64.3, 37.4, 63.5, 101.0, 141.0, 75.2,…
$ `Sr [µg/g] (Leachate)` <dbl> 767, 681, 690, 552, 527, 528, 551, 482, 391…
$ `Ca [µg/g] (Leachate)` <dbl> 188951.9, 163260.4, 162188.7, 125937.6, 124…
$ `Al [µg/g] (Leachate)` <dbl> 10612.7, 11428.4, 5463.0, 3261.5, 2121.7, 1…
$ `Fe [µg/g] (Leachate)` <dbl> 5935.0, 6814.3, 2465.7, 3936.8, 189.6, 7711…
$ `Ba [µg/g] (Residue)` <dbl> 171.0, 198.0, 251.0, 290.0, 315.0, 259.0, 3…
$ `Sr [µg/g] (Residue)` <dbl> 66.6, 71.3, 90.3, 99.8, 106.0, 90.7, 108.0,…
$ `Ca [µg/g] (Residue)` <dbl> 2315.5, 2369.6, 3007.7, 3447.9, 3713.2, 331…
$ `Al [µg/g] (Residue)` <dbl> 29262.5, 35561.4, 45862.4, 52485.6, 55083.0…
$ `Fe [µg/g] (Residue)` <dbl> 14834.8, 18301.4, 24534.5, 30745.3, 28532.2…Better Column Names
we can remove capitalization, spaces, and strange characters from the column names with the function clean_names() from the Janitor Package.
Better Column Names
Rows: 128
Columns: 19
$ event <chr> "SL73BC", "SL73BC", "SL73BC", "SL73BC", "SL73BC…
$ latitude <dbl> 39.6617, 39.6617, 39.6617, 39.6617, 39.6617, 39…
$ longitude <dbl> 24.5117, 24.5117, 24.5117, 24.5117, 24.5117, 24…
$ elevation_m <dbl> -339, -339, -339, -339, -339, -339, -339, -339,…
$ sample_label_barite_sr <chr> "SL73-1", "SL73-2", "SL73-3", "SL73-4", "SL73-5…
$ samp_type <chr> "non-S1", "non-S1", "non-S1", "S1b", "S1b", "S1…
$ depth_m <dbl> 0.0045, 0.0465, 0.0665, 0.1215, 0.1765, 0.1915,…
$ age_ka_bp <dbl> 1.66, 3.13, 4.13, 5.75, 7.30, 7.78, 8.65, 9.94,…
$ ca_co3_percent <dbl> 61.5, 55.1, 53.0, 43.4, 41.8, 42.3, 42.6, 39.1,…
$ ba_mg_g_leachate <dbl> 72.6, 64.3, 37.4, 63.5, 101.0, 141.0, 75.2, 99.…
$ sr_mg_g_leachate <dbl> 767, 681, 690, 552, 527, 528, 551, 482, 391, 70…
$ ca_mg_g_leachate <dbl> 188951.9, 163260.4, 162188.7, 125937.6, 124733.…
$ al_mg_g_leachate <dbl> 10612.7, 11428.4, 5463.0, 3261.5, 2121.7, 12740…
$ fe_mg_g_leachate <dbl> 5935.0, 6814.3, 2465.7, 3936.8, 189.6, 7711.3, …
$ ba_mg_g_residue <dbl> 171.0, 198.0, 251.0, 290.0, 315.0, 259.0, 310.0…
$ sr_mg_g_residue <dbl> 66.6, 71.3, 90.3, 99.8, 106.0, 90.7, 108.0, 96.…
$ ca_mg_g_residue <dbl> 2315.5, 2369.6, 3007.7, 3447.9, 3713.2, 3316.7,…
$ al_mg_g_residue <dbl> 29262.5, 35561.4, 45862.4, 52485.6, 55083.0, 44…
$ fe_mg_g_residue <dbl> 14834.8, 18301.4, 24534.5, 30745.3, 28532.2, 22…Better Column Names
Fixed Variables in Front
Which column is a fixed variable?
I’m not sure if ca_co3_percent is a measured variable, and if it belongs to another informational unit.
Besides that, the fixed variables are already in front.
Pivot Longer
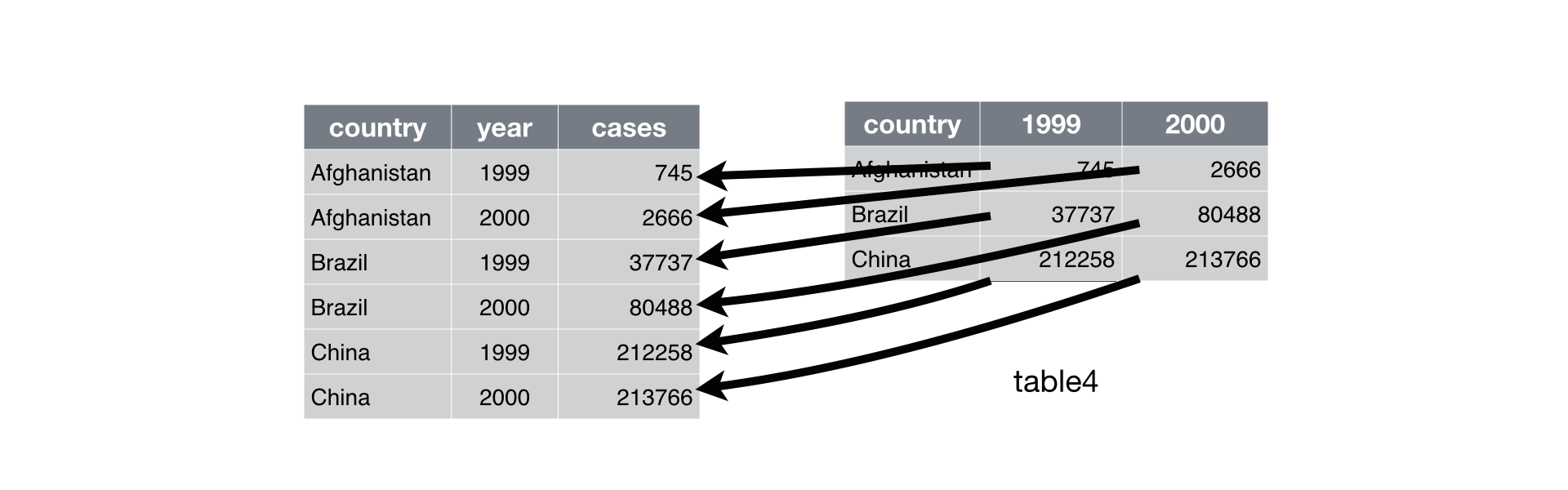
Source : R4DS - Tidy Data
No Values in Column Names
No Values in Column Names
When we pivot data we move them from a wide to a long format and vice-versa.
Rows: 1,280
Columns: 11
$ event <chr> "SL73BC", "SL73BC", "SL73BC", "SL73BC", "SL73BC…
$ latitude <dbl> 39.6617, 39.6617, 39.6617, 39.6617, 39.6617, 39…
$ longitude <dbl> 24.5117, 24.5117, 24.5117, 24.5117, 24.5117, 24…
$ elevation_m <dbl> -339, -339, -339, -339, -339, -339, -339, -339,…
$ sample_label_barite_sr <chr> "SL73-1", "SL73-1", "SL73-1", "SL73-1", "SL73-1…
$ samp_type <chr> "non-S1", "non-S1", "non-S1", "non-S1", "non-S1…
$ depth_m <dbl> 0.0045, 0.0045, 0.0045, 0.0045, 0.0045, 0.0045,…
$ age_ka_bp <dbl> 1.66, 1.66, 1.66, 1.66, 1.66, 1.66, 1.66, 1.66,…
$ ca_co3_percent <dbl> 61.5, 61.5, 61.5, 61.5, 61.5, 61.5, 61.5, 61.5,…
$ element <chr> "ba_ppm_leachate", "sr_ppm_leachate", "ca_ppm_l…
$ concentration <dbl> 72.6, 767.0, 188951.9, 10612.7, 5935.0, 171.0, …One Value Per Cell
Now it’s clear that element contains more than one value.
For example: ba_ppm_leachate is not a single values and could be split into:
- element: ba.
- unit: **ppm*.
- fraction: leachate.
Let’s split this column at the **_** and reconstitute it in a tidy way
One Value Per Cell
One Value Per Cell
Rows: 1,280
Columns: 13
$ event <chr> "SL73BC", "SL73BC", "SL73BC", "SL73BC", "SL73BC…
$ latitude <dbl> 39.6617, 39.6617, 39.6617, 39.6617, 39.6617, 39…
$ longitude <dbl> 24.5117, 24.5117, 24.5117, 24.5117, 24.5117, 24…
$ elevation_m <dbl> -339, -339, -339, -339, -339, -339, -339, -339,…
$ sample_label_barite_sr <chr> "SL73-1", "SL73-1", "SL73-1", "SL73-1", "SL73-1…
$ samp_type <chr> "non-S1", "non-S1", "non-S1", "non-S1", "non-S1…
$ depth_m <dbl> 0.0045, 0.0045, 0.0045, 0.0045, 0.0045, 0.0045,…
$ age_ka_bp <dbl> 1.66, 1.66, 1.66, 1.66, 1.66, 1.66, 1.66, 1.66,…
$ ca_co3_percent <dbl> 61.5, 61.5, 61.5, 61.5, 61.5, 61.5, 61.5, 61.5,…
$ element <chr> "ba", "sr", "ca", "al", "fe", "ba", "sr", "ca",…
$ unit <chr> "ppm", "ppm", "ppm", "ppm", "ppm", "ppm", "ppm"…
$ fraction <chr> "leachate", "leachate", "leachate", "leachate",…
$ concentration <dbl> 72.6, 767.0, 188951.9, 10612.7, 5935.0, 171.0, …Data can take many shapes
Data can take many shapes
Rows: 256
Columns: 16
$ event <chr> "SL73BC", "SL73BC", "SL73BC", "SL73BC", "SL73BC…
$ latitude <dbl> 39.6617, 39.6617, 39.6617, 39.6617, 39.6617, 39…
$ longitude <dbl> 24.5117, 24.5117, 24.5117, 24.5117, 24.5117, 24…
$ elevation_m <dbl> -339, -339, -339, -339, -339, -339, -339, -339,…
$ sample_label_barite_sr <chr> "SL73-1", "SL73-1", "SL73-2", "SL73-2", "SL73-3…
$ samp_type <chr> "non-S1", "non-S1", "non-S1", "non-S1", "non-S1…
$ depth_m <dbl> 0.0045, 0.0045, 0.0465, 0.0465, 0.0665, 0.0665,…
$ age_ka_bp <dbl> 1.66, 1.66, 3.13, 3.13, 4.13, 4.13, 5.75, 5.75,…
$ ca_co3_percent <dbl> 61.5, 61.5, 55.1, 55.1, 53.0, 53.0, 43.4, 43.4,…
$ unit <chr> "ppm", "ppm", "ppm", "ppm", "ppm", "ppm", "ppm"…
$ fraction <chr> "leachate", "residue", "leachate", "residue", "…
$ ba <dbl> 72.6, 171.0, 64.3, 198.0, 37.4, 251.0, 63.5, 29…
$ sr <dbl> 767.0, 66.6, 681.0, 71.3, 690.0, 90.3, 552.0, 9…
$ ca <dbl> 188951.9, 2315.5, 163260.4, 2369.6, 162188.7, 3…
$ al <dbl> 10612.7, 29262.5, 11428.4, 35561.4, 5463.0, 458…
$ fe <dbl> 5935.0, 14834.8, 6814.3, 18301.4, 2465.7, 24534…Pivot Wider
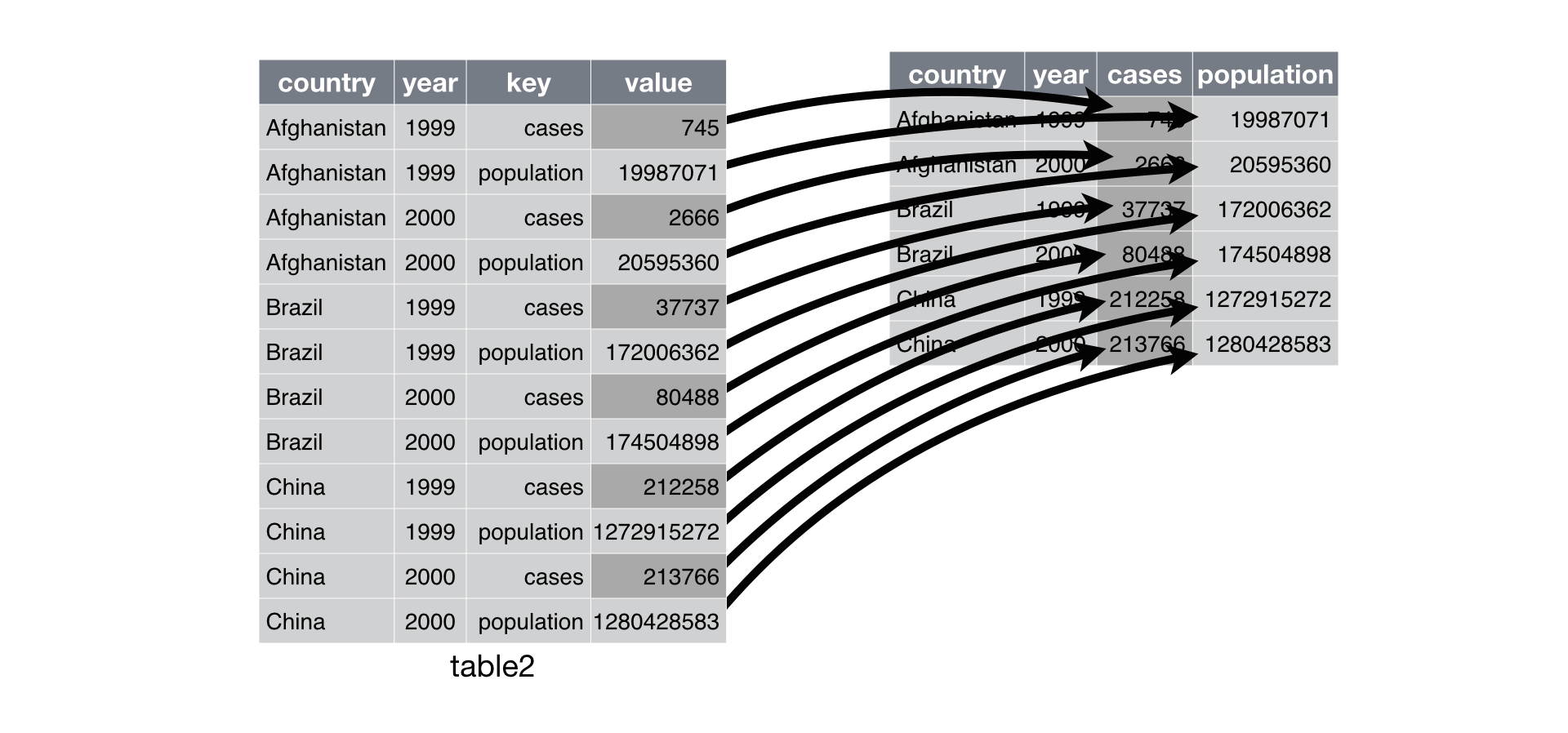
Source : R4DS - Tidy Data
Exercise
Tidy the iris dataset:
# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosa
# ℹ 140 more rowsExercise
Tidy last week’s schedule: